on
Exploring Apache Iceberg with Spark

March 2025 update: use latest Iceberg release 1.8.1
Apache Iceberg is a new table format for storing large and slow moving tabular data on cloud data lakes like S3 or Cloud Storage.
It was developed at Netflix and was then incubated at the Apache Foundation.
It has been a top level project of the Apache Foundation since 2021 and it has reached version 1.0.0 in November 2022.
It brings a new level of abstraction on Parquet format which has now become industry standard for storing datasets in data lakes.
This new level of abstraction aims to solve common issues encountered when working with Parquet format:
- Atomic, consistent and fast updates (e.g. issues when writing Parquet on S3 with Spark I have described in S3A committers article)
- Schema evolution without needs to rewrite entire dataset (new column, column rename, etc.)
- Efficient reads that only read needed partitions
Estimated reading time: 10 minutes
Table of contents
Local Setup
Data and Environment
I am going to use a subset a Covid 19 vaccination data provided by Our World in Data at this address: https://github.com/owid/covid-19-data.
I setup a local S3-like object storage with MinIO on my local machine:
$ minio server ~/Downloads/minio-data --console-address :9001
Automatically configured API requests per node based on available memory on the system: 155
Finished loading IAM sub-system (took 0.0s of 0.0s to load data).
Status: 1 Online, 0 Offline.
API: http://127.0.0.1:9000
RootUser: minioadmin
RootPass: minioadmin
Console: http://127.0.0.1:9001
RootUser: minioadmin
RootPass: minioadmin
Then I create a bucket warehouse on MinIO console available at http://127.0.0.1:9001. This is where data written with Iceberg will be stored.
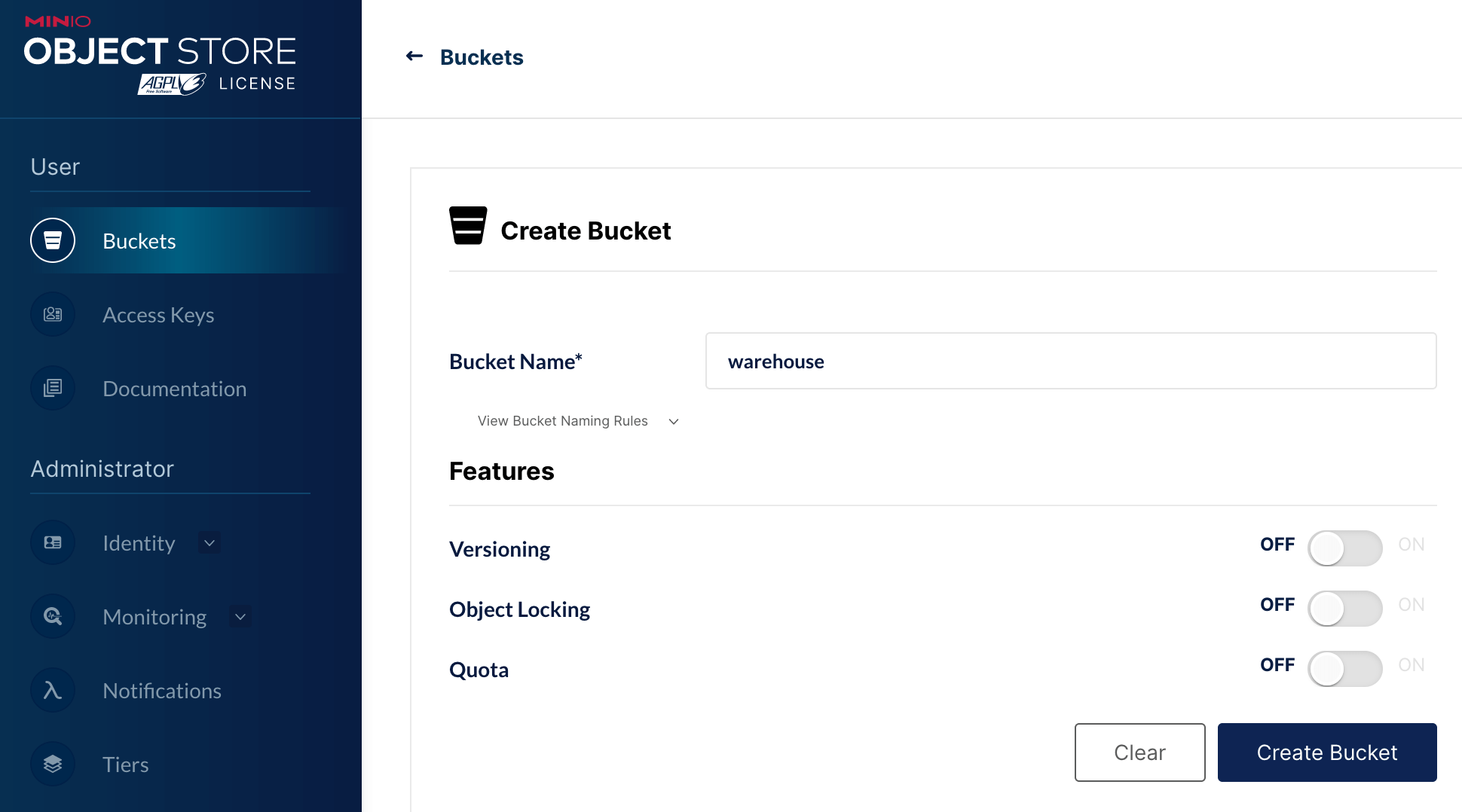
To ensure data consistency in case of concurrent reads and writes, Iceberg tracks data and tables using a database. In my test I use a PostgreSQL database for that purpose. I start a server on default port 5432 and I create a database iceberg_db.
I am going to use Spark framework with Scala for this test. I am familiar with it and it is now the most advanced integration with Iceberg format according to documentation. Iceberg has integrations with other engines such as Apache Flink, Trino (Presto) or Dremio.
Dependencies
I declare versions of Scala, Spark, Iceberg, AWS-SDK and PostgreSQL connector in my Maven pom.xml properties:
<properties>
<scala-major.version>2.13</scala-major.version>
<scala.exact.version>2.13.16</scala.exact.version>
<spark-major.version>3.4</spark-major.version>
<spark-exact.version>3.4.4</spark-exact.version>
<iceberg.version>1.8.1</iceberg.version>
<aws.version>2.31.1</aws.version>
<postgresql.version>42.7.5</postgresql.version>
</properties>
Then I declare dependencies:
<dependencies>
<dependency>
<groupId>org.scala-lang</groupId>
<artifactId>scala-library</artifactId>
<version>${scala.exact.version}</version>
</dependency>
<!-- Spark dependencies -->
<dependency>
<groupId>org.apache.spark</groupId>
<artifactId>spark-core_${scala-major.version}</artifactId>
<version>${spark-exact.version}</version>
</dependency>
<dependency>
<groupId>org.apache.spark</groupId>
<artifactId>spark-sql_${scala-major.version}</artifactId>
<version>${spark-exact.version}</version>
</dependency>
<dependency>
<groupId>org.apache.spark</groupId>
<artifactId>spark-hive_${scala-major.version}</artifactId>
<version>${spark-exact.version}</version>
</dependency>
<!-- Iceberg dependencies -->
<dependency>
<groupId>org.apache.iceberg</groupId>
<artifactId>iceberg-hive-metastore</artifactId>
<version>${iceberg.version}</version>
</dependency>
<dependency>
<groupId>org.apache.iceberg</groupId>
<artifactId>iceberg-spark-runtime-${spark-major.version}_${scala-major.version}</artifactId>
<version>${iceberg.version}</version>
</dependency>
<!-- AWS dependencies to communicate with MinIO warehouse -->
<dependency>
<groupId>software.amazon.awssdk</groupId>
<artifactId>url-connection-client</artifactId>
<version>${aws.version}</version>
</dependency>
<dependency>
<groupId>software.amazon.awssdk</groupId>
<!-- bundle contains SDK clients for all services (S3, KMS, Glue, etc.) -->
<artifactId>bundle</artifactId>
<version>${aws.version}</version>
</dependency>
<!-- PostgreSQL connector to communicate with tracking database -->
<dependency>
<groupId>org.postgresql</groupId>
<artifactId>postgresql</artifactId>
<version>${postgresql.version}</version>
</dependency>
</dependencies>
Spark Job
We need to define configuration in Spark Job according to our test environment based on MinIO and PostgreSQL.
import org.apache.spark.sql.SparkSession
import org.apache.spark.{SparkConf, SparkContext}
object IcebergWithSpark {
def main(args: Array[String]): Unit = {
// Set Minio credentials in Java system properties
System.setProperty("aws.accessKeyId", "minioadmin")
System.setProperty("aws.secretAccessKey", "minioadmin")
val conf = new SparkConf()
.setAppName("Apache Iceberg with Spark")
.setMaster("local[4]")
.setAll(Seq(
// Add Iceberg SQL extensions like UPDATE or DELETE in Spark
("spark.sql.extensions", "org.apache.iceberg.spark.extensions.IcebergSparkSessionExtensions"),
// Register my_iceberg_catalog
("spark.sql.catalog.my_iceberg_catalog", "org.apache.iceberg.spark.SparkCatalog"),
// my_iceberg_catalog tables are tracked in PostgreSQL database
("spark.sql.catalog.my_iceberg_catalog.catalog-impl", "org.apache.iceberg.jdbc.JdbcCatalog"),
("spark.sql.catalog.my_iceberg_catalog.uri", "jdbc:postgresql://127.0.0.1:5432/iceberg_db"),
("spark.sql.catalog.my_iceberg_catalog.jdbc.user", "postgres"),
("spark.sql.catalog.my_iceberg_catalog.jdbc.password", "postgres"),
// my_iceberg_catalog tables contents are stored in MinIO
("spark.sql.catalog.my_iceberg_catalog.io-impl", "org.apache.iceberg.aws.s3.S3FileIO"),
("spark.sql.catalog.my_iceberg_catalog.s3.endpoint", "http://127.0.0.1:9000"),
("spark.sql.catalog.my_iceberg_catalog.warehouse", "s3://warehouse"),
))
val sc = new SparkContext(conf)
val spark = SparkSession.builder.config(sc.getConf).getOrCreate()
// ready to work with Iceberg catalog through Spark session
}
}
Some notes:
- Other catalog implementations exist like GlueCatalog that track tables inside a AWS Glue database or DynamoDbCatalog in AWS DynamoDB.
- Catalog IO implementation S3FileIO is Iceberg implementation that can write and read data with s3 protocol. I configure it to communicate with MinIO endpoint http://127.0.0.1:9000. It requires aws-sdk s3 client to be in your classpath.
- I have set MinIO credentials programatically in Java system properties but S3FileIO relies on AWS default credentials provider which can search credentials at different places: https://docs.aws.amazon.com/sdk-for-java/latest/developer-guide/credentials.html#credentials-chain
Write Data
Once I have configured Iceberg catalog in Spark configuration we can start creating a table and add data to it.
You can interract with Iceberg in Spark are possible either with Spark-SQL or with DataFrameWriterV2 API.
I would recommend to use Spark-SQL for table management like creating a table, dropping it or changing its schema. And use DataFrameWriter API when writing or updating data in table.
import org.apache.spark.sql.SparkSession
import org.apache.spark.sql.functions.{col, to_date}
def createTable(spark: SparkSession) = {
// create table with schema according to vaccinations data
spark.sql("""
CREATE TABLE IF NOT EXISTS my_iceberg_catalog.db.vaccinations (
location string,
date date,
vaccine string,
source_url string,
total_vaccinations int,
people_vaccinated int,
people_fully_vaccinated int,
total_boosters int
) USING iceberg PARTITIONED BY (location, date)"""
)
}
def writeData(spark: SparkSession) = {
val vaccinations: DataFrame = spark.read
.option("header", "true")
.option("inferSchema", "true")
.csv("~/Downloads/covid19-vaccinations-country-data/*.csv")
vaccinations
.withColumn("date", to_date(col("date")))
.sortWithinPartitions("location", "date")
.writeTo("my_iceberg_catalog.db.vaccinations")
.append()
}
Iceberg requires to sort the data according to table partitioning before write in the table. Here I am using
.sortWithinPartitions("location", "date")on my Dataframe before writing it.
Results in MinIO and PostgreSQL
In MinIO under warehouse/db/vaccinations/ we now have 2 directories: data/ and metadata/.
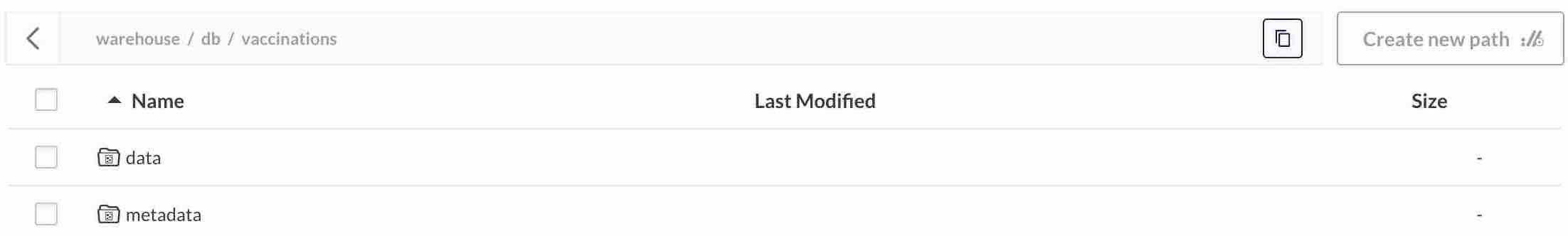
data/ contains data appended in table, it is like what we could had written by saving our covid vaccination data in Parquet with data.write.parquet(...).
metadata/ contains manifest files which store informations about {added/updated/deleted} data under data/ directory. Iceberg will use these manifests to represent table after each modification known as a snapshot. In the end table evolution is a sequence of all manifests added since its creation:
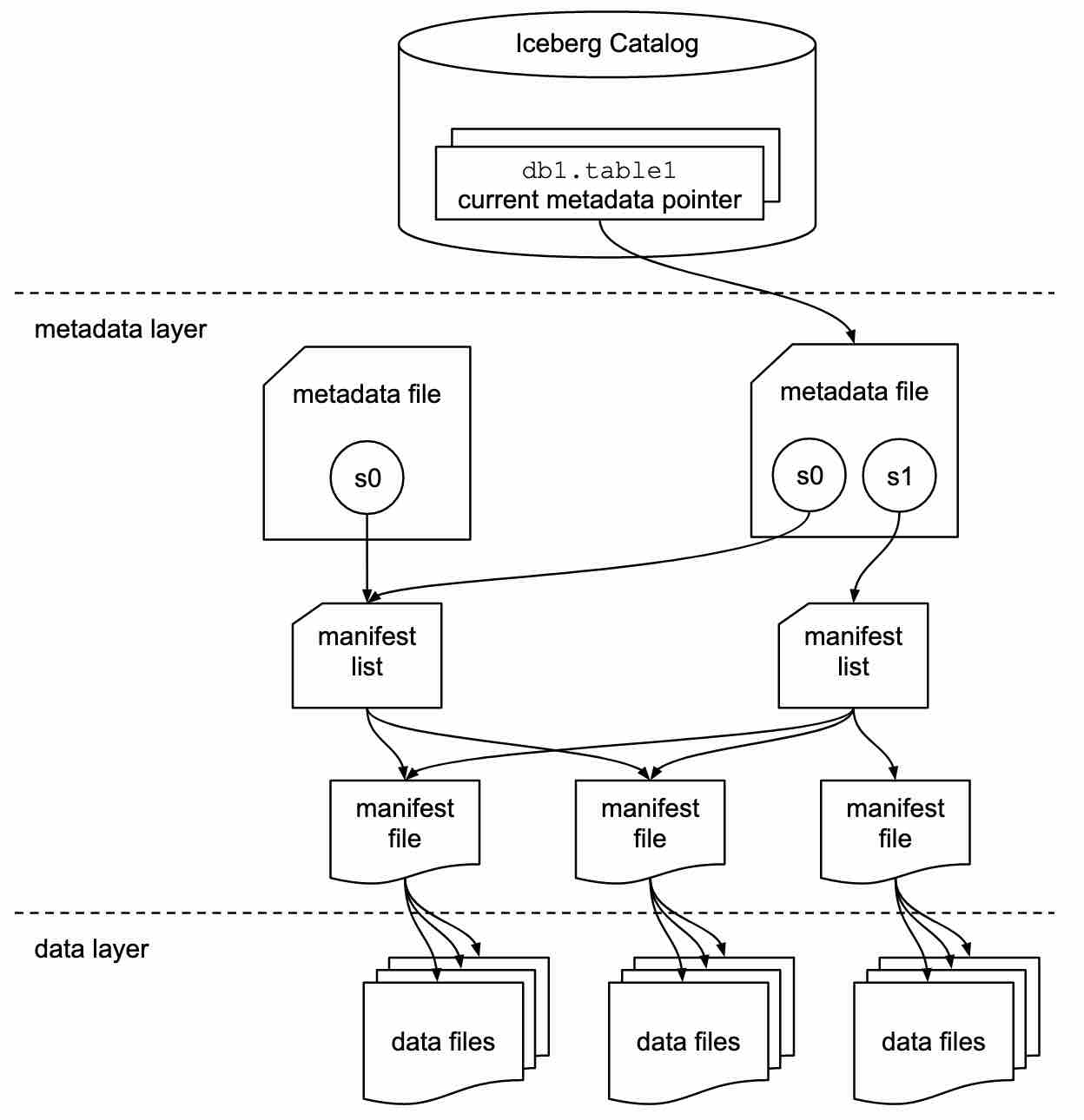 Source: https://iceberg.apache.org/spec
Source: https://iceberg.apache.org/spec
Inside PostgreSQL table:
$ psql -h 127.0.0.1 -d iceberg_db -U postgres -W
Password:
iceberg_db=# \x
Expanded display is on.
iceberg_db=# SELECT * FROM iceberg_tables;
-[ RECORD 1 ]--------------+-------------------------------------------------------------------------------------------------
catalog_name | my_iceberg_catalog
table_namespace | db
table_name | vaccinations
metadata_location | s3://warehouse/db/vaccinations/metadata/00001-f13c501a-f0ad-42e9-8748-6cda27150c08.metadata.json
previous_metadata_location | s3://warehouse/db/vaccinations/metadata/00000-c60c1051-8564-46bb-abaf-1937b841d9a4.metadata.json
We can see current metadata file is 00001-... which corresponds to data insertion. Previous manifest was 00000- which was table creation.
Read Data
We can read Iceberg data with spark.table():
val vaccinations = spark.table("my_iceberg_catalog.db.vaccinations")
vaccinations.limit(5).show()
+--------+----------+---------------+--------------------+------------------+-----------------+-----------------------+--------------+
|location| date| vaccine| source_url|total_vaccinations|people_vaccinated|people_fully_vaccinated|total_boosters|
+--------+----------+---------------+--------------------+------------------+-----------------+-----------------------+--------------+
| Belgium|2020-12-28|Pfizer/BioNTech|https://epistat.w...| 340| 340| 0| 0|
| Belgium|2020-12-29|Pfizer/BioNTech|https://epistat.w...| 380| 379| 11| 0|
| Belgium|2020-12-30|Pfizer/BioNTech|https://epistat.w...| 911| 909| 21| 0|
| Belgium|2020-12-31|Pfizer/BioNTech|https://epistat.w...| 959| 957| 21| 0|
| Belgium|2021-01-01|Pfizer/BioNTech|https://epistat.w...| 976| 974| 21| 0|
+--------+----------+---------------+--------------------+------------------+-----------------+-----------------------+--------------+
Hidden partitionning
This is one cool feature of Iceberg. Thanks to separation between physical and logical layout we don’t need to filter data according to how it is partitioned to prevent full data scans and slow queries.
In my vaccinations table data I have partitioned data by location and by date: location=Brazil/date=2020-12-29/*.parquet
I can query table by omitting location and still have a fast read query, Iceberg will only select necessary partitions in locations that have this date:
val vaccinations = spark.table("my_iceberg_catalog.db.vaccinations")
vaccinations.filter(col("date") === "2021-05-01").select("location").distinct().show()
+--------------------+
| location|
+--------------------+
| China|
| United States|
| Denmark|
....
Partitioning can evolve without need to rewrite historical data. For example we want to change the 2nd part of vaccinations table partitionning from actual date (date=2020-12-29) to only keep the year and month parts (date=2020-12).
spark.sql("ALTER TABLE my_iceberg_catalog.db.vaccinations DROP PARTITION FIELD date")
spark.sql("ALTER TABLE my_iceberg_catalog.db.vaccinations ADD PARTITION FIELD months(date)")
// new data added in table will now be partitioned by month
Iceberg will be able to split following query:
spark.table("my_iceberg_catalog.db.vaccinations")
.filter(col("date") === "2021-05-01")
It will select {location}/{date} before and {location}/month({date}) after partition change occured.
Overwrite Data
We can use overwritePartitions() instead of append() to overwrite data:
newVaccinationsStats
.writeTo("my_iceberg_catalog.db.vaccinations")
.overwritePartitions()
Under the hood overwritten data is not deleted in data/ directory in warehouse. Instead new data is added and a new manifest that links those new files is created. When Iceberg finishes overwritting it commits changes in PostgreSQL table. This way, if we have any concurrent reads while updating table, they don’t see potential incorrect data.
Schema evolution
Iceberg supports a wide range of possible schema evolutions. They are listed here: https://iceberg.apache.org/docs/latest/spark-ddl/#alter-table.
In my example, I am going to add a new column in my table which represents the percentage of fully vaccinated people:
spark.sql("""
ALTER TABLE my_iceberg_catalog.db.vaccinations
ADD COLUMN people_fully_vaccinated_percentage double
""")
With standard Parquet I would have needed to add mergeSchema option to have this new column for historical data. With Iceberg I am able to see this new column even if data already present in table didn’t have it. It will be null by default.
I verify correct behavior by adding data of Spain which was not yet in table. I compute this new column before writing it:
val spainVaccinations = spark.read....
spainVaccinations
.withColumn("date", to_date(col("date")))
.withColumn(
"people_fully_vaccinated_percentage",
(col("people_fully_vaccinated") / col("people_vaccinated")) * 100
)
.sortWithinPartitions("location", "date")
.writeTo("my_iceberg_catalog.db.vaccinations")
.append()
And then when I read it:
spark.table("my_iceberg_catalog.db.vaccinations")
.filter(col("location") === "Spain")
.select("location", "date", "people_fully_vaccinated_percentage")
.show(1)
+--------+----------+----------------------------------+
|location| date|people_fully_vaccinated_percentage|
+--------+----------+----------------------------------+
| Spain|2021-01-31| 28.60003723921561|
+--------+----------+----------------------------------+
Conclusion
Apache Iceberg looks very promising. It is still in young phase, but the developer experience is pleasant.
Existing documentation, examples and integrations help you to dive into this new table format.
Some of the largest companies like Netflix, Apple or Adobe are using it at scale in production.
Iceberg main competitor is Delta Lake format from Databricks which aims to solve same issues when working with Parquet format.
Links
- Code, data and environment setup I have used are available here: https://github.com/romibuzi/iceberg-spark-demo
- Iceberg official documentation: https://iceberg.apache.org/docs/latest/
- Iceberg: a fast table format for S3 at DataWorks Summit 2018 by Ryan Blue co-creator of Iceberg: https://youtu.be/nWwQMlrjhy0
heading image source: spalla67 at https://pixabay.com